cath-cluster
A simple way to complete-linkage cluster arbitrary data.
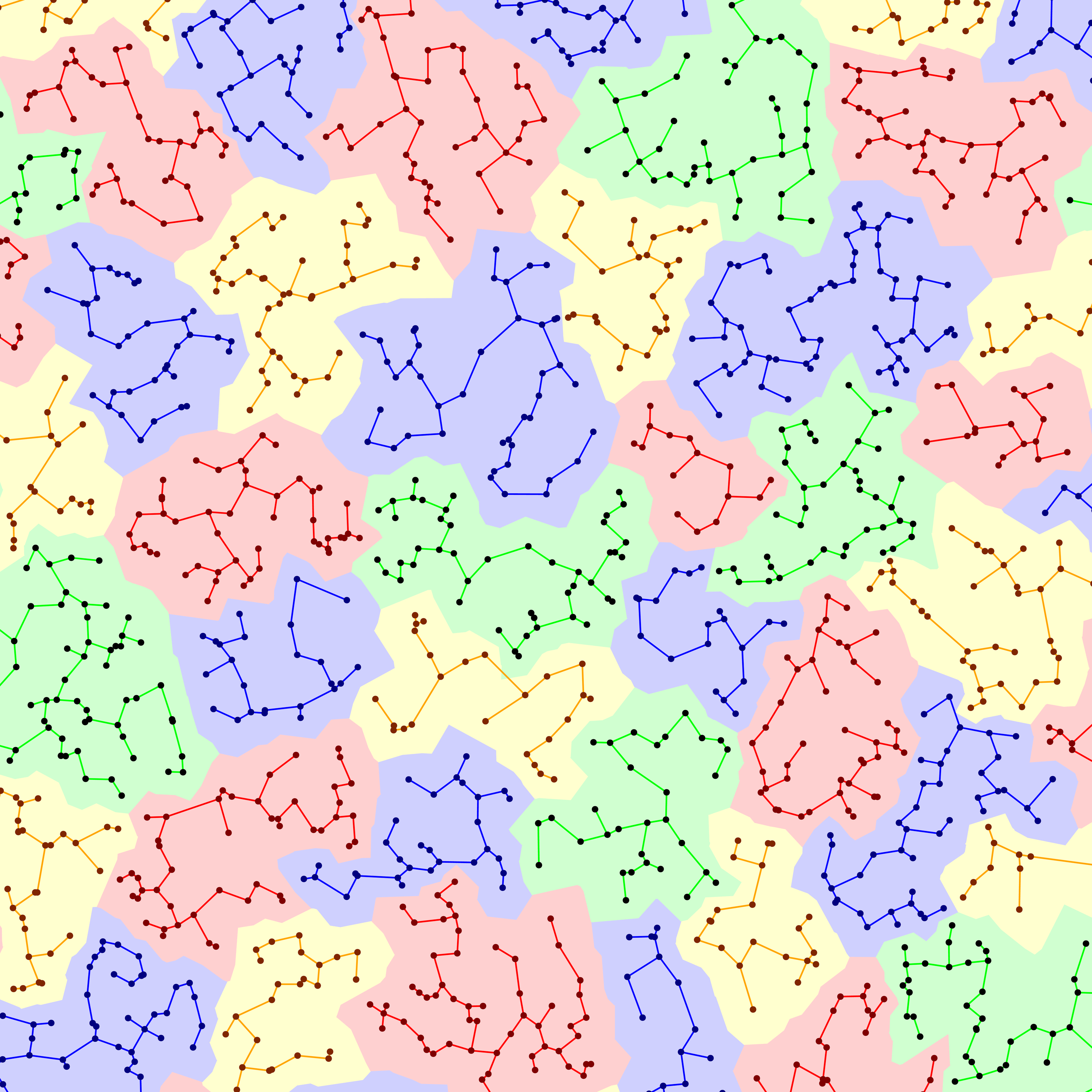
Above: A complete-linkage clustering of randomly generated points based on the distances between them
Features
- Fast
- Simple
Usage
The current full --help usage information is:
Usage: cath-cluster --link_dirn <dirn> [options] <input_file>
Cluster items based on the links between them.
When <input_file> is -, the links are read from standard input.
The clustering is complete-linkage.
Miscellaneous:
-h [ --help ] Output help message
-v [ --version ] Output version information
Input:
--link_dirn <dirn> Interpret each link value as <dirn>, one of:
DISTANCE - A higher value means the corresponding two entries are more distant
STRENGTH - A higher value means the corresponding tow entries are more connected
--column_idx <colnum> (=3) Parse the link values (distances/strengths) from column number <colnum>
Must be ≥ 3 because columns 1 and 2 must contain the IDs
--names-infile <file> [RECOMMENDED] Read names and sorting scores from file <file> (or '-' for stdin)
Clustering:
--levels <levels> Cluster at levels <levels>, which is ordered values separated by commas (eg 35,60,95,100)
Output:
--clusters-to-file <file> Write the clustering to file <file> (or '-' for stdout)
--merges-to-file <file> Write the ordered list of merges to file <file> (or '-' for stdout)
--clust-spans-to-file <file> Write links that form spanning trees for each cluster to file <file> (or '-' for stdout)
--reps-to-file <file> Write the list of representatives to file <file> (or '-' for stdout)
Links input format: `id1 id2 other columns afterwards`
...where --column_idx can be used to specify the column that contains the values
Names input format: `id score`
...where score is used to sort such that lower-scored entries appear earlier
Please tell us your cath-tools bugs/suggestions : https://github.com/UCLOrengoGroup/cath-tools/issues/new
Feedback
Please tell us about your cath-tools bugs/suggestions here.